Artificial gene synthesis, or gene synthesis, alludes to a gathering of strategies that are utilized in engineered biology to build and amass genes from nucleotides anew. Dissimilar to DNA synthesis in living cells, artificial gene synthesis doesn't need layout DNA, permitting essentially any DNA grouping to be orchestrated in the research center.
It contains two fundamental advances, the first is strong stage DNA synthesis, now and then known as DNA printing. This produces oligonucleotide sections that are generally under 200 base sets. The subsequent advance at that point includes associating these oligonucleotide sections utilizing different DNA gathering techniques. Since artificial gene synthesis doesn't need format DNA, it is hypothetically conceivable to make totally engineered DNA particles unbounded on the nucleotide grouping or size.
Synthesis of the primary complete gene, a yeast tRNA, was shown by Har Gobind Khorana and colleagues in 1972. Synthesis of the main peptide-and protein-coding genes was acted in the labs of Herbert Boyer and Alexander Markham, respectively. More as of late, artificial gene synthesis techniques have been fostered that will permit the gathering of whole chromosomes and genomes.
Also read: DNA Modifications in Humans
The primarily manufactured yeast chromosome was blended in 2014, and whole practical bacterial chromosomes have likewise been synthesized. also, artificial gene synthesis could, later on, utilize novel nucleobase sets.
The compound synthesis of DNA oligonucleotides and their gathering into synthons, genes, circuits, and surprisingly whole genomes by gene synthesis techniques has become an empowering innovation for current atomic biology and empowers the plan, construct, test, learn, and rehash cycle supporting developments in manufactured biology.
In this point of view, we momentarily survey the methods and innovations that empower the synthesis of DNA oligonucleotides and their get together into bigger DNA builds with an emphasis on late progressions that have tried to diminish synthesis cost and increment arrangement constancy. The improvement of cheaper techniques to create excellent manufactured DNA will take into account the investigation of bigger biological theories by bringing down the expense of utilization and help to close the DNA read–compose cost hole.
It has been said that the twentieth century was the "hundred years of the particle" in which disclosures on the physical and synthetic properties of the components prompted leap forwards like nuclear energy, clinical diagnostics, PCs, and the computer chip to give some examples.
Also read: Why AIDS is an Incurable Disease?
These advances dramatically affected our lifestyle and aided shape the guarantee and conceivable outcomes of science and innovation. In the early piece of the 21st century, we are seeing what could almost certainly get known as the "hundred years of DNA." Like the score to life's perplexing ensemble, DNA gives the plan to biological capacity. Advances in the course of the most recent couple of a very long time in both perusing and composing (synthesis) DNA groupings have rolled out stamped improvements in our capacity to comprehend and design biological frameworks.
These headways have prompted the advancement of pivotal innovations for the plan, gathering, and control of DNA encoded genes, materials, circuits, and metabolic pathways, which are considering an always more noteworthy control of biological frameworks and surprisingly whole organic entities.
Because of cutting-edge sequencing (NGS) innovations equipped for generating an expected 15 petal bases of succession information each year around the world (Schatz and Phillippy 2012), the current metagenomics period has prompted the growth of biological grouping archives with DNA arrangements disconnected from each organic entity and climate possible. Related enhancements in bioinformatics procedures and programming permit specialists to acquire, examine, and control these DNA arrangements in manners simpler than any time in recent memory.
The steadily expanding accessibility of biological grouping data from all parts of the "tree of life" has developed our comprehension of biological frameworks and the interrelated idea of living beings at the genetic level. NGS advancements have prompted a more profound comprehension of human infections, the microbiome, and the genetic variety of living beings in our current circumstances.
This succession blast is additionally taking into account the extension of logical teaches, for example, metabolic designing and engineered biology as specialists try to utilize novel groupings in the control of biological frameworks for human-centric methods. Furthermore, this abundance of data is prompting the advancement of an assortment of diagnostics and therapeutics, which will add to the drawn-out progress of human wellbeing.
This biological arrangement information gold mine has been helped by a surge of developments in instrumentation and methods for generating sequencing information with high loyalty, expanded throughput, and diminished expense, which has added to making NGS a go-to innovation for some applications in biology.
The capacity to generate enormous DNA arrangement informational indexes has permitted analysts to make and test huge perused biological speculations utilizing NGS advancements that were unrealistic before their turn of events. Past dissecting biological frameworks at the DNA grouping level, the need to build DNA from planned arrangements has driven the equal advancement of strategies and instrumentation to create manufactured DNA at scale to empower the testing of designed biological parts.
Also read: Theory of Evolution by Charles Darwin
DNA perusing by NGS when joined with present-day DNA synthesis innovations structure the two essential advances driving manufactured biology endeavors and will in the long run ingrain the consistency and dependability to designed biological frameworks that synthetic designing has brought to substance frameworks. This being said, our capacity to grouping DNA is at present better compared to our capacity to incorporate DNA again, albeit new advancements are assisting with shutting the hole.
Engineered biology is arising as a significant control with the possibility to affect various scholastic and mechanical applications including the formation of novel therapeutics, materials, biosensing, and producing abilities. Even though our present comprehension of biological frameworks is tremendous, it is still a long way from complete. Adjusting exogenous DNA groupings and the practical parts they encode that have normally advanced inside an organization of interrelated successions stays a rule challenge of designing biology.
As of now, the designing of biological frameworks requires a weighty portion of experimental experimentation to assess novel compounds, articulation frameworks, and pathways for the ideal capacity. Normally, this cycle is cultivated by first planning an ideal engineered biological circuit or pathway utilizing PC-supported plan apparatuses.
Then, the subsequent build DNA is separated into more modest covering pieces (regularly 200–1500-bp fragments) that are simpler to orchestrate. These DNA parts are then incorporated from a bunch of covering single-abandoned oligonucleotides in-house (or by business merchants). The subsequent covering synthons are then gathered into bigger parts of DNA and cloned into an articulation vector and the grouping of the subsequent development is then checked. The arrangement confirmed develops are then changed into a cell and examined for work. Contingent upon the outcomes, changes to the build configuration can be made and further emphasis of the test cycle rehashed.
This plan, fabricate, test, learn, and rehash measure has become the foundation of engineering biology, which thusly has put a premium on computerized cycles and techniques that can abbreviate the improvement cycle and increment throughput.
One of the qualities that make biological frameworks alluring from a designing viewpoint is the way that biological capacities are encoded to a huge part in DNA. Accordingly, gross rearrangements of biological designing can be decreased to the plan, creation, and testing of DNA groupings. As scientists try to design biological frameworks with novel DNA groupings, the requirement for custom manufactured DNA successions has developed.
This is especially obvious when the groupings to be designed are gotten from metagenomic arrangements in which no creature might be accessible from which to segregate the DNA through different strategies. The synthesis of engineered DNA is regularly alluded to generically as "gene synthesis," which explicitly is the synthesis of gene-length bits of DNA (250–2000 bp) straightforwardly from single-abandoned manufactured DNA oligonucleotides.
Sadly, while the expense of sequencing has diminished steeply after some time, the expense of gene synthesis and oligonucleotide synthesis, in general, has not kept speed, albeit mechanical advancement and market influences are dynamically bringing down the expense of engineered DNA. The expense of gene synthesis is regularly straightforwardly attached to the expense of oligonucleotide synthesis from which the genes are made. The expense of oligonucleotide synthesis has not diminished obviously in over 10 years, generally going from $0.05 to $0.17 per base contingent upon the synthesis scale, the length of the oligonucleotide, and the provider (Kosuri and Church 2014).
Customarily, this expense floor has been brought through to the creation of gene synthesis items. These manufactured genes presently range in cost from $0.10 to $0.30 per bp ($100–$300 for a 1-kb gene) from conventional business providers, even though organizations abusing fresher cheaper synthesis techniques are beginning to cut the expense down.
Bringing down the expense of gene synthesis would empower the generation of bigger informational collections by making the expense of gene development more affordable, implying that more builds could be generated for a similar expense venture. This would permit scientists the capacity to test a more noteworthy measure of the planning scene.
With a more prominent comprehension of what does constantly not work in a given development plan, a bunch of general plan decisions could be made that would improve the accomplishment of the plan interaction and ultimately abbreviate the plan fabricate test–learn measure. Since the significant expense of gene synthesis is the reagents that are required for oligonucleotide synthesis, moves toward that decrease reagent utilization, improve the vigor and exactness of the gene get together cycle, and empower expanded throughput have become important instruments for propelling the use of manufactured DNA by taking into consideration lower cost of utilization.
In this viewpoint, a concise outline of the strategies and innovations that have added to the creation of engineered DNA using gene synthesis techniques will be featured just as a portion of the difficulties that have must be defeated to lessen the expense, increment the throughput, and guarantee the devotion of manufactured DNA.
OLIGONUCLEOTIDE SYNTHESIS
Current all over again gene synthesis strategies can follow their beginnings to the mid-1960s and Gobind Khorana and associates endeavors to unravel the genetic code and to reconstitute biological capacity artificially. These early endeavors zeroed in on artificially orchestrating and ligating together 17 oligonucleotides, which encoded the gene for a 77-nucleotide alanine tRNA utilizing engineered sciences still in their early stages in an exertion that required over 5 years to finish.
Within 7 years of this work, the primary detailed manufactured gene encoding the 14-amino-corrosive chemical, somatostatin, was communicated in recombinant Escherichia coli.
Ensuing enhancements to oligonucleotide synthesis sciences and procedures in the mid-1980s prompted the advancement of strong stage phosphoramidite sciences whose strength and devotion took into consideration computerized strategies to be created to empower adaptable oligonucleotide synthesis that is utilized right up 'til the present time for the business synthesis of oligonucleotides.
Manufactured oligonucleotides for gene synthesis applications are generally combined utilizing varieties of the phosphoramidite science strategies either on customary segment-based synthesizers or microarray-based synthesizers that will be examined in the accompanying segments.
Section-BASED OLIGONUCLEOTIDE SYNTHESIS
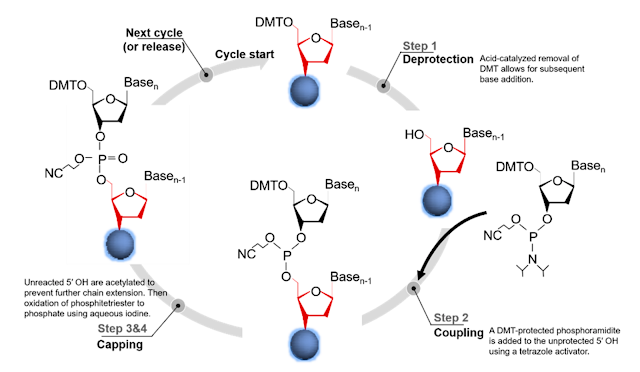
The regularly utilized phosphoramidite synthesis science comprises of a four-venture chain lengthening cycle that adds one base for each cycle onto a developing oligonucleotide anchor connected to a strong help lattice. In the initial step, a dimethoxytrityl (DMT)- ensured nucleoside phosphoramidite that is appended to a strong help is deprotected by the expansion of trichloroacetic corrosive.
This actuates the help connected phosphoramidite for chain lengthening with the following phosphoramidite monomer. In the subsequent advance, the following base in the arrangement is included the type of a DMT-ensured phosphoramidite and is coupled to the 5′-hydroxyl gathering of the past nucleoside phosphoramidite in the grouping framing a phosphite triester.
Third, any unreacted 5′-hydroxyl bunches are covered by acylation to deliver any unextended arrangements inactive in ensuing rounds of the chain extension cycle and along these lines diminishing erasure mistakes in the completed oligonucleotide groupings. In the fourth step, the phosphite triester linkage between the monomers is changed over to a phosphate linkage using oxidation with an iodine answer to produce a cyanoethyl-secured phosphate spine. The synthesis cycle at that point rehashes for the following base in the arrangement using the expulsion of the 5′-terminal DMT ensuring bunch.
After the ideal succession has been orchestrated from the 3′ to 5′, the oligonucleotide is synthetically cut from the strong synthesis support and the securing bunches on the bases and the spine are eliminated. This cycle is profoundly agreeable to mechanization and structures the reason for oligonucleotide synthesizers, which can integrate 96–1536 unmistakable oligonucleotides all the while. These synthesizers commonly orchestrate oligonucleotides at scales going from 10 to 1000 nmol with an expense running somewhere in the range of $0.05 and $0.17 per base. These oligonucleotides can generally be combined up to ∼100 nt with blunder rates at or beneath 0.5%. Section-based coupling yields are generally very productive (normally >99% per coupling); notwithstanding, the yield of full-length oligonucleotides regularly declines with expanding oligonucleotide length.
Yields of full-length oligonucleotides can be influenced by false depurination of the blended oligomer, particularly at adenosines during the corrosive deprotection steps of the synthesis cycle, and turns out to be particularly dangerous for longer oligonucleotide groupings. These false abasic locales can lessen the yields of full-length oligonucleotides by advancing the cleavage of the oligonucleotide phosphate spine during the expulsion of the excess nucleobase and spine securing bunches following the last synthesis cycle. An extra decrease of synthesis quality is brought about by the presentation of synthesis-related blunders into the integrated successions, basically as single-base erasures.
The fundamental wellspring of this sort of blunder is the fragmented expulsion of the DMT securing bunch or the consolidated shortcomings of the coupling and covering ventures during the synthesis cycle.
Albeit complete evacuation of synthesis-related blunders will probably be unimaginable in light of under 100% response efficiencies in a stage astute various response synthesis, late enhancements in the synthesis cycle and the sciences utilized will probably prompt increases in the length and nature of oligonucleotides orchestrated not long from now.
For gene synthesis applications, because each oligonucleotide orchestrated is made on singular synthesis sections the oligonucleotides important to collect a gene should be added together into a gathering pool post-synthesis, which characteristically diminishes the oligonucleotide synthesis pool intricacy because just the oligonucleotides required for a given synthesis are remembered for the gathering combination.
MICROARRAY-BASED OLIGONUCLEOTIDE SYNTHESIS
An option in contrast to customary section-based oligonucleotide synthesis has risen up out of the utilization of microarray oligonucleotide synthesis. These innovations, which were initially created for delivering oligonucleotide microarrays for analytic applications, have arisen as a promising minimal expense option in contrast to segment-based oligonucleotide synthesis.
Early forms of these synthesizers combined the oligonucleotides appended to a computer chip surface utilizing a change of the phosphoramidite synthesis measure. Affymetrix was one of the early pioneers in this field and grew light-initiated sciences to control the spatially isolated synthesis of oligonucleotides on the chip surface utilizing standard veil-based photolithography methods to specifically deprotect unique photolabile nucleoside phosphoramidites.
Organizations, for example, NimbleGen and LC Sciences further worked on the light-coordinated synthesis systems by disposing of the photolithography veils by utilizing programmable micromirror gadgets to absolutely control the light-based sciences. Then again, ink-fly printing-based advances created by Agilent and semiconductor-based chip advances by CombiMatrix (CustomArray) consider the utilization of standard phosphoramidites and reagents without the requirement for costly micromirror regulators or photomasking.
Specifically, these two synthesis innovations at present give the best blend of cost, oligonucleotide synthesis length, and exactness, which has prompted their utilization in a few ongoing gene synthesis applications. Utilizing microarray-based synthesizers, oligonucleotides are regularly combined at femtomolar synthesis scales, that is, ordinarily two to four significant degrees lower than that utilized in segment-based synthesis.
Also read: Can we Make Mars Habitable for Life
This lower synthesis scale prompts an attending decline in the number of reagents required for the synthesis. The decrease of reagent used in the synthesis cycle prompts drastically lower costs. Notwithstanding the diminished synthesis scale, chip-based oligonucleotide synthesizers offer degrees of multiplexing unrealistic on customary segment-based synthesizers that takes into account a more noteworthy number of exceptional oligonucleotide successions to be combined in a given synthesis run.
With conceivable component densities during the many thousands for every chip and the ability for certain synthesizers to integrate various chips all the while, the capacity of microarray-based synthesis techniques far overwhelms the limit of even the biggest section-based instruments. The joined diminished synthesis scale and huge multiplexing ability implies that oligonucleotide synthesis costs from exhibit-based stages can go from $0.00001 to $0.0001 per nucleotide relying upon the stage, oligonucleotide length, and synthesis scale.
When contrasted and the $0.05–$0.10 cost per base feasible for section orchestrated oligonucleotides, the allure of chip-integrated oligonucleotides for gene synthesis applications comes to be unmistakable.
Although exhibit-based stages offer unrivaled synthesis abilities as far as multiplexing and cost there are a few difficulties with utilizing these stages for making oligonucleotides for gene synthesis applications. One issue is that planar exhibit integrated oligonucleotides will, in general, be moderately bad quality, which prompts more synthesis-related mistakes when contrasted and section orchestrated oligonucleotides.
Regardless of this, late upgrades in chip plan and refinements to the synthesis cycle have prompted an expansion like exhibit integrated oligonucleotides. One such wellspring of diminished synthesis quality is the depurination of oligonucleotide successions on chips, which can happen suddenly in light of delayed openness of the incipient oligonucleotides to the deprotecting reagents during the synthesis cycle.
Enhancements to the response conditions and the progression of reagents to the chip during the synthesis cycle have been appeared to adequately improve the nature of the oligonucleotides integrated on chips by diminishing depurination and expanding the general synthesis yields, which thus empowers the synthesis of oligonucleotides up to 200 nts long.
Another wellspring of diminished synthesis quality with chip created oligonucleotides is a result of "edge impacts," which is brought about by misalignment of the reagent beads with the chose chip to include or erroneous particular deprotection of the appropriate synthesis highlights brought about by ill-advised reagent sequestration or light pillar floats in synthesizers utilizing light-initiated synthesis sciences.
Edge impacts diminish oligo quality by lessening the particular control of the synthesis responses in nearby oligonucleotides, which can prompt replacement or erasure blunders in oligonucleotides integrated on neighboring highlights. Work to diminish edge impacts during the synthesis cycle has shown that refinements to the chip configuration can improve the loyalty of cluster incorporated oligonucleotides to match segment orchestrated oligonucleotides.
Proceeded with progress in exhibit plan and refinements to synthesis reagents and cycles utilized will prompt routine hearty synthesis of top-notch long oligonucleotides from clusters, which will advance their utilization as the go-to hotspot for minimal expense oligonucleotides for gene synthesis applications.
GENE SYNTHESIS FROM OLIGONUCLEOTIDES
Since DNA is a polymer comprised of four distinctive nucleotide monomers, gene synthesis and DNA gathering techniques are as a result a type of progressive polymer synthesis. For manufactured DNA, individual phosphoramidite monomers are consolidated together to make individual oligonucleotides 60–100 nt long.
To work with the gathering of an engineered twofold abandoned DNA (synthon) from single-abandoned oligonucleotides, neighboring oligonucleotides are intended to contain covering arrangements between the oligonucleotides encoded on the restricting strands of the DNA duplex and are collected together during the gene synthesis cycle to make twofold abandoned synthons from 200 to 2000 bp long.
Succession-checked synthons would then be able to be collected together by various strategies to make bigger manufactured DNA builds encoding whole metabolic pathways or genomes.
Early oligonucleotide gathering strategies depended on the ligation of contiguous oligonucleotides through the utilization of a DNA ligase.
At first, the ligation of nearby oligonucleotides was done successively, yet as gathering strategies and the nature of the engineered oligonucleotides improved and the utilization of thermostable ligases were utilized to improve the severity of the get-together response the one-pot gathering of DNA synthons through ligation got conceivable.
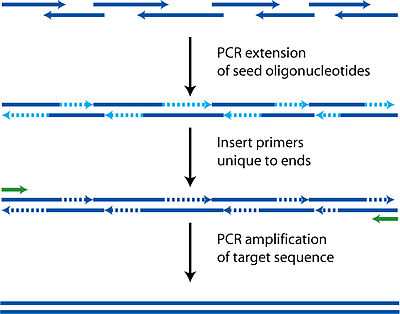
Not long after the revelation of the polymerase chain response (PCR) during the 1980s sans ligation techniques for utilizing manufactured single-abandoned oligonucleotides were created to make totally engineered bits of DNA without the requirement for a normally inferred DNA format.
Since this time, many various strategies have been made to amass twofold abandoned DNA from single-abandoned oligonucleotides through PCR-like techniques. A rundown of a considerable lot of these techniques has been inspected in the writing and won't be surveyed broadly here. These strategies utilize single-abandoned engineered oligonucleotides with integral covering groupings between adjoining oligonucleotides to collect twofold abandoned DNA synthons utilizing a thermostable DNA polymerase and PCR.
The solitary contrasts between the heap of PCR-based DNA gathering strategies are in how the substituent oligonucleotides are intended to be amassed together and the response conditions under which they are collected, the finished result of these techniques is something similar, a twofold abandoned DNA (dsDNA) synthon.
Of these gathering techniques, varieties of the polymerase cycle get together (PCA) strategy is by a long shot the most ordinarily used to amass DNA synthons from oligonucleotides. Utilizing PCA, a dsDNA grouping is isolated into oligonucleotide successions (regularly 60–80 nt), which encode the two strands of the DNA duplex with covers between neighboring oligonucleotides that reach from 15 to 25 nt long. Ordinarily, contiguous oligonucleotides are planned with holes between the forward and the converse covering locales of the get-together oligonucleotides to decrease the measure of oligonucleotide synthesis needed to orchestrate a given succession.
Once planned and blended, the substituent oligonucleotides for a gathering are pooled together in equimolar fixations and cycled in a one-pot "gathering" response in which adjoining oligonucleotides are arbitrarily stretched out in a nonexponential way by a DNA polymerase to create a combination of oligonucleotide expansion results of different lengths. This combination is then utilized as a layout to seed a second PCR response, in which the ideal "full-length" item is enhanced from the gathering blend within the sight of an overabundance of the peripheral get-together preliminaries.
Notwithstanding ligation and PCA-based gathering strategies, Gibson and partners, as a component of their spearheading work in engineered genomics, have created both in vitro and in vivo one-pot, single-step techniques for at the same time gathering DNA synthons from oligonucleotides and cloning them straightforwardly into plasmids. In some variety, these three gathering techniques are utilized in general business and scholarly gene synthesis applications are answered to date.
GENE SYNTHESIS FROM ARRAY-DERIVED OLIGONUCLEOTIDE POOLS
Throughout the most recent decade, the longing for less expensive wellsprings of manufactured DNA has filled an interest in cluster combined DNA as an approach to deliver oligonucleotides modestly. Past early exhibitions of gene synthesis from cluster-created oligonucleotides, late advancements have made generous upgrades to the quality, productivity, and versatility of microarray-based oligonucleotide synthesis and gene gathering.
Also read: Neuroscience in Robotic Technologies
To utilize microarray-blended DNA for gene synthesis, a few obstacles have must be defeated to make this wellspring of DNA an appealing option to oligonucleotides combined by regular segment-based methodologies. A portion of the properties that make cluster combined oligonucleotides appealing as a minimal expense wellspring of DNA for gene synthesis present difficulties for really utilizing oligonucleotides blended in this manner for gene synthesis.
The decreased synthesis size of cluster-created oligonucleotides is appealing from an expense point of view due to the associative decrease in reagent utilization expected to integrate a given DNA succession. Be that as it may, the measure of DNA created (fmol) is generally considerably less than what is required (pmol) to dependably amass them into bigger synthons. Also, the enormous multiplexing abilities of cluster-based oligonucleotide synthesis imply that a huge number of interesting oligonucleotide arrangements can be integrated at the same time on the exhibit surface.
Nonetheless, the grouping intricacy of the oligonucleotide pools delivered on the exhibit can prompt issues with amassing them into bigger builds because of the obstruction between oligonucleotides that contain even humble succession homology. Last, as referenced already, exhibit combined oligonucleotides are in general lower quality as contrasted and their segment incorporated partners, which shows itself as succession mistakes in the engineered DNA collected from them. Thus, in general, the utilization of exhibit incorporated oligonucleotides requires the utilization of some kind of blunder decrease procedure to lessen the work needed to acquire succession checked develops.
To defeat these, several unique techniques have been created to build the compelling oligonucleotide pool fixation to guarantee solid get together and reproducibility. Tian et al. (2004) tended to the focus issue by including a typical preparing arrangement followed by a scratching endonuclease acknowledgment site on the 3′-finish of each oligonucleotide blended on the cluster.
The exhibit blended oligonucleotides fill in as the layout strand for a groundwork augmentation response utilizing the basic preparing arrangement consolidated into the cluster-connected oligonucleotide pool. Following groundwork expansion, a scratching endonuclease remembered for the response combination eliminates the basic preliminary groupings to deliver a solitary abandoned DNA oligonucleotide that is integral to each oligonucleotide orchestrated on the exhibit. This cycle enhances the cluster combined oligonucleotide pool to build the centralization of the oligonucleotides to levels reasonable for gene synthesis.
Also read: Most Powerful Bionic Eye Ever
In this technique, because the exhibit blended oligonucleotides stay appended to the cluster, spatial control of oligonucleotide get together can happen on-chip. To misuse this element and to improve the multiplexing abilities of chip-based oligonucleotide synthesis, Quan et al. (2011) utilized a custom ink-stream synthesizer that orchestrated oligonucleotides on particular chips. Inside these particular chips, miniature compartmentalized gathering wells can be planned into the synthesis exhibit with the oligonucleotides important to collect an exceptional synthon, incorporated, enhanced, and amassed inside individual microwells.
Utilizing such a strategy viably decreases the succession intricacy of the confined oligonucleotide pool, which thus builds the vigor of gathering for gene synthesis while additionally considering synthesis multiplexing that can happen in each of the numerous microwells planned into the chip.
Another technique previously presented by Kosuri et al. (2010) tackles the oligonucleotide focus and pool intricacy issues by including "bar-coded" preparing arrangements into each of the oligonucleotides incorporated on the exhibit. In this technique, the oligonucleotides needed to combine an ideal synthon are flanked on the two finishes by sets of predefined groundwork groupings.
The peripheral preparing successions consider the enhancement of the cluster blended oligonucleotide pool, which has been separated from the chip surface after synthesis. This progression considers one bunch of novel preliminaries to intensify the whole oligonucleotide pool with the end goal that a bit of the pool can be utilized for resulting get-together or chronicled purposes. Inboard of the pool enhancement groundworks is a bunch of flanking preliminary restricting arrangements that consider specific intensification of get-together sub-pools from the synthesis pool.
Every special synthon would be given a novel arrangement of flanking sub-pool preliminary groupings to such an extent that the oligonucleotides important to gather any given synthon would be specifically intensified from the synthesis pool for ensuing get together. Following enhancement of the sub-pool oligonucleotides, the preparing groupings are taken out by limitation processing of the intensified dsDNA oligonucleotides utilizing a special chemical acknowledgment succession flanking the get-together oligonucleotide arrangement.
Also read: What is Antimatter and How it is made?
This oligonucleotide configuration plot at the same time tackles the oligonucleotide focus and pool intricacy issues related to cluster inferred oligonucleotides and doesn't need specific chips or exhibit synthesizers. This strategy is additionally profoundly manageable to computerization as each sub-pool can be gathered in special wells in a multiwell plate design utilizing PCA get-together strategies.
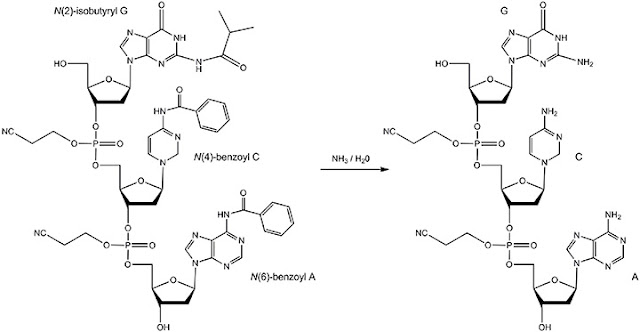
CORRECTION AND SEQUENCE VALIDATION
One of the central difficulties in the synthesis and gathering of manufactured DNA from unpurified oligonucleotides is managing arrangement mistakes brought into the groupings by synthesis results and the get-together interaction itself. Following the gathering, an engineered DNA can be considered as a populace of successions containing a combination of right and inaccurate arrangements.
Customarily, the manufactured DNA would be cloned into a vector and individual clones segregated and sequenced by Sanger sequencing. In any case, the likelihood of some random succession containing mistake increments with the length of DNA, which thus implies that more clones should be sequenced to acquire a totally right clone. This technique is tedious and the arrangement work required adds fundamentally to the expense of the DNA being delivered.
To diminish the number of clones important to get the right succession, some post-synthesis strategy should be applied to the gathered DNA to strainer the right arrangements from inaccurate ones. Therefore, techniques have been fostered that estimated the blunder adjusting measure that phones use to keep up arrangement devotion during DNA replication. In general, most techniques for eliminating DNA grouping mistakes from manufactured DNA start with the development of a DNA heteroduplex.
This is finished by warming up the manufactured DNA to denature and disassociate the strands followed by cooling the example to reanneal the strands together. For some random situation inside a DNA succession, the greater part of the duplexes in the engineered combination will contain the right base at that situation with mistakes sprinkled all through the populace.
Since grouping blunders happen arbitrarily inside a gathered DNA arrangement this denaturation and reannealing measure prompt the development of heteroduplexes DNA at places that contain mistakes. Places that contain transformations inside these heteroduplexes can be followed up on by proteins, which explicitly perceive succession changes in DNA.
One such gathering of techniques depends on the grouping bungle acknowledgment abilities of the MutS protein to explicitly tie to arrangement befuddles in manufactured DNA duplexes. Specific restricting of MutS to mistake containing DNA can be utilized to sifter blunder-free successions from those that contain mistakes.
These techniques generally immobilize MutS to a strong framework material and afterward sanitize segment bound (blunder containing) DNA arrangements from unbound material (mistake diminished). Blunder containing heteroduplex DNA can likewise be sieved utilizing catalysts that perceive and cut the DNA duplex at the site of the base confuse.
The utilization of endonuclease proteins (or compound mixed drinks), which perceive and cut DNA heteroduplexes at the locales of bungles, has been demonstrated to be exceptionally successful at decreasing synthesis-related blunders in manufactured genes taking into consideration time and material reserve funds to such an extent that at times the treated genes can be utilized straightforwardly in practical measures without cloning and grouping check.
These strategies are a generally clear and financially savvy approach for sieving mistake containing successions from manufactured DNA and can diminish the noticed blunder rate from 1/100 bases to 1/10,000 bases.
An option in contrast to compound interceded blunder remedy methods is the direct sequencing of oligonucleotides and collected synthons utilizing NextGen sequencing strategies.
Albeit more costly for every succession premise than catalyst-based blunder rectification strategies, NextGen sequencing procedures offer gigantic multiplexing abilities in which a great many arrangement peruses can be taken on various oligonucleotides or synthons all the while.
In one such application, Matzas et al. (2010) utilized Roche 454 sequencing joined with a globule picking robot to specifically eliminate oligonucleotide successions (connected to dots) from the sequencing exhibit that showed wonderful groupings that were then utilized for gene synthesis. Kim et al. (2012) utilized arbitrary succession labels to stamp singular engineered arrangements in a 454 sequencing response and utilized these labels to specifically enhance right groupings from the sequenced pool.
In a comparable methodology, Schwartz et al. (2012) utilized Illumina sequencing scanner tags to "dial-out" right arrangements using specific enhancement of the grouping checked bar-coded successions. As of late, they have refined this way to deal with improve the multiplexing limit of microarray-orchestrated DNA to pairwise gather 2271 131–250 bp synthons from a solitary oligonucleotide pool and utilized bar-coded groundworks to specifically detach grouping checked people.
These strategies highlight manners by which NextGen advances can be utilized to improve the nature of manufactured DNA. Furthermore, because arrangement-based methodologies assess assortments of individual particles of DNA, they are reasonable for the grouping confirmation of manufactured DNA, which may contain areas of succession degeneracy like libraries for coordinated development.
Also read: Can We Create an Artificial Sun?
At present, every one of these techniques is restricted by the grouping read-length capacities of the NextGen instrument utilized and by sequencing-related mistakes. Be that as it may, as NextGen instrumentation and methods keep on improving, these restrictions will turn out to be less critical and consider the exact confirmation of longer bits of engineered DNA.
MAKE IT BIGGER: SYNTHESIS OF LONGER DNA ASSEMBLIES
Ongoing progressions in oligonucleotide synthesis and strategies for gene synthesis have generally centered around decreasing expenses, expanding throughput, and the nature of manufactured DNA.
Attending progress in gathering DNA into longer and longer pieces have prompted techniques to develop enormous protein edifices, whole metabolic pathways, and surprisingly whole genomes artificially.
Strategies to amass synthons have been created to gather synthons and bigger DNA gatherings both in vitro and in vivo. Although limitation protein-based cloning procedures have been the de rigueur decision for controlling DNA builds for years and years and were the premise of early BioBrick and comparable gathering strategies, the need to work on the cloning/gathering measure while diminishing the limits on arrangement configuration has prompted the advancement of scar-less limitation compound free cloning and get together methods.
Creaseless get together and cloning strategies accessible to the cutting edge DNA jockey incorporate Gibson gathering, Golden Gate gathering, succession and ligation-free cloning (SLIC), ligation cycling response, paper-cut get together, yeast gets together, and round polymerase expansion cloning (CPEC), to give some examples. Which DNA gathering strategy to utilize is to a great extent a question of decision, and numerous methodologies are frequently applied in equal. Of these techniques, Gibson gathering is likely the most ordinarily used to collect different bits of DNA together into bigger builds.
This strategy utilizes a one-pot isothermal procedure, which utilizes a compound combination containing a thermostable DNA polymerase, DNA ligase, and exonuclease to bite back, strengthen and fix nearby covering DNA successions to collect the ideal development.
Ongoing developments in the plan of novel covering successions to coordinate the get-together cycle have additionally extended the use of the Gibson gathering strategy for the combinatorial gathering of huge DNA arrangements.
One of the signs of engineered biology is the utilization of objective plan standards to the plan and get together of biological segments; notwithstanding, because it is regularly hard to know deduced how well a given DNA build will work once brought into a cell, it is frequently important to attempt a few renditions of the developers to discover which one will work best.
Thusly, a more prominent accentuation on the secluded plan of DNA parts empowers the get together of a more noteworthy assortment of possible develops through blend and-match combinatorial gathering of DNA segments. As well as improving on the general get-together cycle, measured plan, and gathering of DNA parts make mechanization of the interaction conceivable, which can decrease the time, work, and cost of making and testing different builds.
A large portion of the previously mentioned get-together techniques can be utilized for the gathering in vitro, and Gibson get-together has been applied to the get-together of DNA sections different kilobases long. In another such model, Gibson gets together was utilized to amass the 16.3-kb mouse mitochondrial genome straightforwardly from 60 mer oligonucleotides.
Effective get-together of much bigger engineered DNA portions can likewise be acted in vivo by utilizing the homologous recombination capacities of the yeast Saccharomyces cerevisiae.
In an illustration of the extraordinary capacity of yeast to amass exogenous DNA into bigger congregations from covering synthons or subassemblies, analysts at the J. Craig Venter Institute have effectively utilized yeast to gather various 0.5–1 Mbp bacterial genomes and surprisingly gathered synthons straightforwardly from covering oligonucleotides.
Each of the previously mentioned get-together methods could be robotized to additional expansion the throughput for developing bigger manufactured DNAs and empower the investigation and testing of enormous biological speculations.
Applications
As DNA printing and DNA get-together techniques have permitted business gene synthesis to turn out to be logically and dramatically less expensive over the past years, artificial gene synthesis addresses an incredible and adaptable designing device for making and planning new DNA groupings and protein capacities.
Other than manufactured biology, different examination regions like those including heterologous gene articulation, antibody advancement, gene treatment, and sub-atomic designing, would profit extraordinarily from having quick and modest strategies to combine DNA to code for proteins and peptides. The techniques utilized for DNA printing and get-together have even empowered the utilization of DNA as a data stockpiling medium.
Incorporating bacterial genomes
On June 28, 2007, a group at the J. Craig Venter Institute distributed an article in Science Express, saying that they had effectively relocated the normal DNA from a Mycoplasma mycoides bacterium into a Mycoplasma capricolum cell, making a bacterium that acted like an M. mycoides.
On Oct 6, 2007, Craig Venter reported in a meeting with UK's The Guardian paper that a similar group had integrated an adjusted adaptation of the single chromosome of Mycoplasma genitalium artificially. The chromosome was adjusted to dispose of all genes which tests in live microbes had demonstrated to be superfluous.
The following arranged advance in this insignificant genome project is to relocate the blended negligible genome into a bacterial cell with its old DNA eliminated; the subsequent bacterium will be called Mycoplasma laboratorium. The following day the Canadian bioethics bunch, ETC Group gave an assertion through their delegate, Pat Mooney, saying Venter's "creation" was "a skeleton on which you could fabricate nearly anything". The blended genome had not yet been relocated into a working cell.
On May 21, 2010, Science detailed that the Venter bunch had effectively combined the genome of the bacterium Mycoplasma mycoides from a PC record and relocated the integrated genome into the current cell of a Mycoplasma capricolum bacterium that had its DNA eliminated.
The "manufactured" bacterium was practical, for example, equipped for reproducing billions of times. The group had initially intended to utilize the M. genitalium bacterium they had recently been working with, yet changed to M. mycoides because the last bacterium develops a lot quicker, which converted into faster experiments.
Venter portrays it as "the first species.... to have its folks be a computer". The changed bacterium is named "Synthia" by ETC. A Venter representative has declined to affirm any forward leap at the hour of this composition.
Manufactured Yeast 2.0
As a component of the Synthetic Yeast 2.0 task, different exploration bunches all throughout the planet have taken part in a venture to combine manufactured yeast genomes, and through this interaction, streamline the genome of the model organic entity Saccharomyces cerevisiae.
The Yeast 2.0 undertaking applied different DNA get-together techniques that have been talked about above, and in March 2014, Jef Boeke of the Langone Medical Center at New York University uncovered that his group had blended chromosome III of S. cerevisiae. The system included supplanting the genes in the first chromosome with manufactured adaptations and the completed engineered chromosome was then incorporated into a yeast cell.
It required planning and making 273,871 base sets of DNA – less than the 316,667 sets in the first chromosome. In March 2017, the synthesis of 6 of the 16 chromosomes had been finished, with the synthesis of the others still ongoing.
Conclusion
The improvement of automatable strong sciences for compound DNA synthesis in the course of the most recent 40 years has added to the progression of our comprehension of biology and has laid the preparation for the anticipated designing of biological frameworks.
Manufactured DNA is fundamental to the improvement of techniques to design biology and when joined with the monstrous measures of grouping information being generated by NGS endeavors will add to the progression of engineered biology toward applications until now incredible.
Until this point in time, there have been a small bunch of moonshot shows like the total synthesis of a whole yeast chromosome, a whole bacterial genome, and the ensuing synthesis of an insignificant bacterial genome, which outline the utilization of manufactured DNA and the abilities to existing gene synthesis techniques to achieve enormous scope engineered biology endeavors.
These models, when joined with various ventures in which engineered DNA has been utilized to assess utilitarian biological parts, make engineered antibodies, and develop engineered genetic circuits and when applied to discount genomic altering, highlight a future where the subtleties of biological capacity can partially be perceived by the plan, synthesis, and test of compatible engineered segments much the same as manufactured medication advancement.
To empower bigger scope designing endeavors and to democratize the utilization of manufactured biology methods and plan standards, ongoing developments have tried to bring down the expense of combining DNA oligonucleotides and their resulting gathering into engineered builds prepared for use.
The utilization of microarray/microfluidic oligonucleotide synthesis methods, when joined with robotized gene synthesis conventions and upgrades in engineered DNA quality, has taken extraordinary steps toward this objective.
Because of market pressures, commercialization of these advancements and the proceeded improvement of other proficient DNA synthesis methods will additionally upgrade the accessibility of progressively modest manufactured DNA.
Albeit the synthesis of generally little synthons (<1 kb) can frequently be direct and achieved at the undergrad level the gathering of bigger parts of engineered DNA can in any case represent a few specialized difficulties to even the most experienced DNA jockey.
Issues made by a serious level of optional construction in the objective succession, also as redundant DNA groupings, can present difficulties to their gathering. Arrangements that are unconsciously poisonous to the host cells utilized for halfway controls of the DNA or examination of manufactured DNA develops can likewise present issues for the fruitful gathering and utilization of some engineered DNA builds. Here too, further upgrades in the engineered build plan and in the DNA get-together interaction will probably empower the effective synthesis of even the most troublesome objective successions in the not-to-removed future.
Which began with an engineered tRNA gene over 40 years prior has prompted the new synthesis of the primary insignificant bacterial genome and profound enthusiasm for the intricacies and difficulties of designing biology. A lot of which actually still needs to be investigated. In this, the hundred years of DNA, the inquiry remains. What is straightaway?









0 Comments
Thanks for your feedback.